Italy Pushes Back as Health Care Workers Shun Covid Vaccines
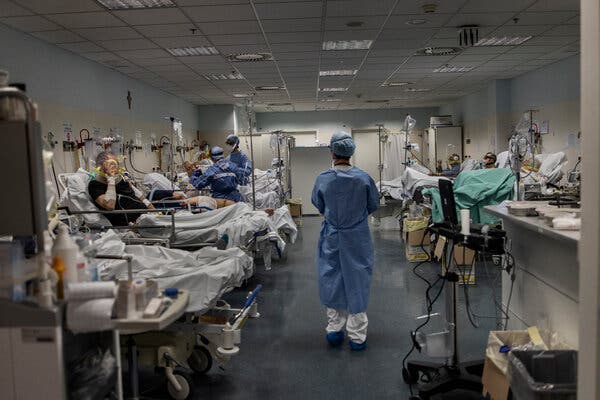
Prime Minister Mario Draghi issued a decree requiring that workers in health care facilities be vaccinated, a move that will test the legal limits of his government’s efforts to stem coronavirus outbreaks.ROME — Giulio Macciò tested negative for the coronavirus and spent weeks receiving treatment for emphysema in a sealed-off hospital under the care of doctors and lung specialists — and a nurse who had refused to be vaccinated. On March 11, he unexpectedly died. A post-mortem swab found that he had contracted the virus, as had 14 other patients and the unvaccinated nurse who spent her shifts in his midst.“It makes no sense that a person whose job is to heal the sick gives them Covid and kills them,” said Mr. Macciò’s son, Massimiliano Macciò, who filed a complaint against the San Martino hospital in the northern Italian city of Genoa. He believes that the nurse, one of an estimated 400 who have refused vaccination against Covid-19 at the hospital, infected his father, who died unvaccinated at 79.As vaccination rollouts build momentum, businesses everywhere are grappling with whether they can require the inoculation of their employees, raising thorny ethical, constitutional and privacy issues around Europe and the United States. But that quandary becomes all the more urgent when the person is your health care worker.In Italy, the original Western front in the war against Covid, a rash of outbreaks in hospitals where medical workers have chosen not to be inoculated has raised fears that their stance is endangering public health. It has also prompted a forceful response from an Italian government that is struggling to get vaccinations on track.On Wednesday, Prime Minister Mario Draghi tested the legal limits of his government’s ability to address the problem by issuing a decree requiring that workers in health care facilities be vaccinated. It also allowed hospital employers to suspend without pay any health care workers who refuse to do so.Some legal analysts have said that requiring Covid-19 inoculation for health workers could violate Italy’s privacy laws, and that firing or forcing any who decline it to take unpaid leave could be unconstitutional because of a specific article that protects people who refuse health treatments.A coronavirus ward at the Papa Giovanni XXIII hospital in Bergamo in March last year.Fabio Bucciarelli for The New York TimesBut recent court rulings have interpreted the law differently, and Mr. Draghi has made clear that for a country that has suffered more than 100,000 Covid deaths, the breach of safety cannot be tolerated.“It is absolutely not OK that unvaccinated workers are in contact with the sick,” he said at a news conference last week while announcing his government’s intention to “intervene” when asked about the reports of unvaccinated health care workers.For much of the pandemic, nurses and doctors stood as national heroes who sacrificed their waking hours, safety and sometimes lives to protect their compatriots. It has shocked Italians that in some major hospitals up to 15 percent of those medical professionals — who were given preference in the vaccination rollout ahead of older people — have shunned inoculation.“It’s really humiliating for the medical and health worker class that you have to force people to vaccinate themselves,” said Roberto Burioni, a virologist at San Raffaele University in Milan.He added that while firing workers is exceedingly difficult in Italy, he hoped the decree will bite into the salaries of any vaccine skeptics, especially considering the large amount of data demonstrating that the vaccines’ efficacy is worth the risk. He also worried that the high number of health professionals refusing to get vaccinated had troubling implications.“Unfortunately there is huge part of doctors who are deeply ignorant,” said Mr. Burioni, who suggested that perhaps “the selection process for bringing people to gain a medical degree and then the medical license is not effective enough.”Transporting coffins in Bergamo last year. Italy has had more than 100,000 Covid-19 deaths.Fabio Bucciarelli for The New York TimesWhile Italy’s populists, including the Five Star Movement and League parties, exploited vaccine skepticism for political gain in recent years, the country is not even considered the most vaccine-skeptic in Europe, a dubious distinction that usually falls to France. Italy also had a fast start in vaccinations at the beginning of the year precisely because the previous government prioritized medical workers.In January, the health minister, Roberto Speranza, said on television that Italy, like its European partners, believed it was better to persuade people to get vaccinated than to require them to. “Those who had to deal with the virus, our health care workers, are even more aware than the others,” he said. “I think willingness will be enough.”But Italy’s vaccination program has hit speed bumps. First, the pharmaceutical company AstraZeneca failed to make good on millions of promised doses. The previous government, led by the Five Star Movement, fell, and Mr. Draghi came in promising to help accelerate vaccinations and the country’s economic recovery.He has pushed for bans on vaccine exports from the European Union to contend with the shortages. He has sought to mobilize new categories of vaccinators and centralize Italy’s response to make up for the failure of some of the country’s hardest-hit regions to inoculate the most vulnerable, older citizens. On Tuesday, Mr. Draghi himself received an AstraZeneca dose after joining a temporary Europe-wide suspension of the vaccine amid concerns about its safety.“It is absolutely not OK that unvaccinated workers are in contact with the sick,” said Prime Minister Mario Draghi, center.Angelo Carconi/EPA, via ShutterstockBut the anti-vax health workers have struck a deep nerve.In a nursing home outside Rome, nearly all of the health care workers chose not to be vaccinated, and a cluster erupted around three workers and 27 out of the 36 older guests. Roberto Agresti, the home’s owner, feared the worst for them. “If we had a law forcing everyone to get vaccinated, the virus would have passed without us even noticing it,” he said.In the southern city of Brindisi, the local health authority has opened disciplinary proceedings against 12 health care workers who expressly refused vaccination. It is also investigating why about 140 health care workers, including doctors, nurses, pediatricians and specialists, declined shots of the Pfizer vaccine.“We don’t want to punish workers — we need them,” said Giuseppe Pasqualone, who leads the local health authority. “But the risk of contagion not only for them but for fragile patients is very high.”Officials at the San Martino hospital, where Mr. Macciò died, said it was not clear whether the unvaccinated nurse was the source of the cluster, but they acknowledged that it was a problem.Salvatore Giuffrida, the director of the hospital, Europe’s fourth largest, said he favored a vaccination requirement because it would also keep medical workers healthy and would strengthen defensive lines as a brutal third wave spreads through northern Italy.“We cannot afford not having them on the job,” he said. “The objective is not to lose soldiers during a war in a nation that complains about not having health care workers.”He estimated that 15 percent of his nursing staff, about 400 nurses, was unvaccinated. Simply removing those nurses from the wards, or redirecting them to switchboards as some have proposed, would be “a cure worse than the disease,” he said, because it would result in the reduction of 250 beds.He and other directors said that Italy’s strict privacy laws kept the hospitals from knowing which doctors and nurses were unvaccinated.Paolo Petralia, the director general of the Lavagna hospital in Chiavari, the site of another outbreak this month, said 90 percent of his doctors were vaccinated, along with about 80 percent of nurses and aides.“They are protected by privacy laws,” he said, citing a recent pronouncement by Italy’s data protection authority that the vaccination status of health workers should be unknown. “But this right exists until it does not limit another person’s right,” Mr. Petralia said.Lining up at a drive-through vaccination center in Milan this month.Alessandro Grassani for The New York TimesSome Italian courts have agreed. In 2017, Italy made some vaccinations compulsory for children, including for measles, and barred the unvaccinated from attending school — a decision backed by Italy’s constitutional court because it also safeguarded public health. In the northern city of Belluno, a court ruled in mid-March that a nursing home that employed several health care workers who chose not to get vaccinated could force them to take paid leave.Mr. Macciò, whose father died in Genoa, said it made no sense that the people entrusted to care for his father were allowed to potentially harm him. He said he had complained to the doctors, who told him their hands were tied because the nurses were protected by privacy rules.But amid Italy’s frustration, and the new decree, something appears to be changing. Mr. Macciò said the police had asked for his help in identifying the nurses he saw when going to pick up his father’s belongings.“I hope some good comes of it,” he said of his father’s death. “These people should change their job.”Emma Bubola
Read more →