India Covid: Patients dying without oxygen amid Delhi surge
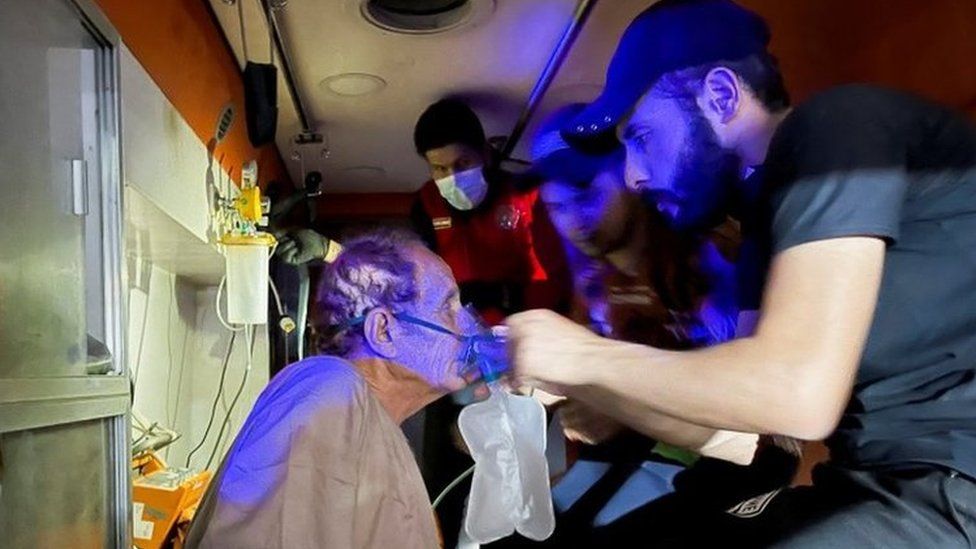
SharecloseShare pageCopy linkAbout sharingFor a fourth day in a row, India has set an unwelcome world record for the number of new coronavirus infections: a further 349,691 cases in the 24 hours to Sunday morning, with another 2,767 lives lost. The capital, Delhi, is one of the worst-hit areas. The BBC’s Vikas Pandey reports from a city whose hospitals are overwhelmed and whose citizens are in desperation. When Ashwin Mittal’s grandmother’s oxygen saturation level dropped a week ago, he started frantically looking for a hospital bed in Delhi. He called everybody he could, but every hospital refused.Her condition deteriorated further on Thursday and he took her to the emergency rooms of several hospitals, but every place was full. They accepted the fate that she was going to die without getting any treatment. But she was gasping for every breath and Ashwin just couldn’t bear it after a while. He took her in his car and went from one hospital to another for several hours until one in north Delhi agreed to take her in the emergency ward for “a few hours”. He was to continue looking for a bed.Ashwin, who has also tested positive for coronavirus, continued his search while battling a high fever and severe body aches. But he couldn’t find a bed, and the hospital continued to keep his grandmother in the emergency ward on compassionate grounds.Deaths climb as India reels from deadly Covid waveViral picture that defines India’s Covid distressWhy second Covid wave is devastating IndiaDoctors there said she needed an ICU and had a good chance of survival. A family friend told me that the hospital was planning to discharge her on Sunday as it was running out of oxygen.”The family is back to where they started and has accepted the fate. They know that if she survives, it will be because of a miracle, not because of any treatment,” the friend said.Miracles are what many families in Delhi are left to rely on. Most hospitals are full and many of them are refusing new admissions owing to the uncertainty over oxygen supply.Oxygen-equipped ambulances are in short supply and it’s becoming difficult for families to transport patients to hospitals even if they find a bed. image copyrightGetty ImagesIndia in crisisDelhi is extending its lockdown for a second weekIndian Prime Minister Narendra Modi has urged people to exercise caution and get vaccinated, saying ” this storm has shaken the nation” The Fortis Escorts Heart Institute, a private hospital in Delhi, said on Sunday it would not admit any more patients because of a shortage of oxygen At least 20 people died on Saturday at the Jaipur Golden Hospital in Delhi because of a lack of oxygenI know a few cases where patients have died because they did not get high-flow oxygen support. Every morning starts with frantic calls from friends, family and colleagues asking for a bed, oxygen cylinders or medicines. The number of people I am able to help is reducing every day as the doctors and officials who could earlier help are no longer available to speak on the phone. Helplines are not working and the vendors who could earlier help have run out of supplies.I go to bed with a sense of defeat every night, but then pick myself up and start in the morning again as more and more people call for help. I can understand their helplessness as I lost a cousin a few days ago in a top hospital in the city. He waited for 18 hours to get a ventilator but the hospital didn’t have any. That’s how Delhi is functioning at the moment. Friends are calling friends; social media is awash with desperate pleas for help.But it’s almost impossible to find a hospital bed here now. Oxygen cylinders and medicines are in short supply. On Saturday, Saroj Hospital and Batra Hospital told families to take their patients away as they were running out of oxygen. The city has been been reporting more than 24,000 cases daily in the past few days. The hospitals are completely overrun, and healthcare workers are exhausted. Some people I know are taking dangerous journeys with their critically ill relatives to other cities which are 300-500km (18-300 miles) away. Sivesh Rana’s brother was in a critical condition but he couldn’t find a bed in Delhi and decided to take him in an ambulance to a city in the neighbouring state of Haryana. But his condition worsened during the journey and the ambulance wasn’t equipped to deal with a critical patient. He died a few hours after arriving at the hospital.Dr A Fathahudeen, who is part of Kerala state’s Covid task force, says the crisis is unprecedented and doctors can’t do much if oxygen supply is not guaranteed.”You need high-pressure liquid oxygen for the smooth functioning of ventilators and bi-pap machines. When the pressure drops, the machines fail to deliver adequate oxygen into the lungs, and the consequences can be fatal,” he says.He adds that oxygen is one of the major treatments to stabilise the patient, allowing doctors time to assess them and plan a future course of treatment. image copyrightGetty ImagesDr Fathahudeen says urgent measures are needed as patients are dying without getting the treatment they require.”The Indian army is one of the finest in the world to build makeshift hospitals and ICU beds within a short span of time. They should be roped in,” he adds.The situation is not very different in other cities, including Pune, Nashik, Lucknow, Bhopal, Indore and Allahabad, as the second Covid wave devastates the country.India reported 349,000 cases on Friday – a record daily spike. It reported 2,767 deaths. But experts say the actual numbers are likely to be much higher.Getting tested has become very difficult in many cities as labs are overrun. As I earlier reported, many people are dying at home because they are not getting hospital beds or they are not being able to get tested for Covid. So, they don’t find a place as a Covid patient in the database managed by different states.Meanwhile, frantic calls continue – each one more heartbreaking than the last. How have you been affected by coronavirus in India? Tell us your story by emailing: haveyoursay@bbc.co.uk.Please include a contact number if you are willing to speak to a BBC journalist. You can also get in touch in the following ways:WhatsApp: +44 7756 165803Tweet: @BBC_HaveYourSayUpload your pictures/video hereOr fill out the form belowPlease read our terms & conditions and privacy policy
If you are reading this page and can’t see the form you will need to visit the mobile version of the BBC website to submit your question or comment or you can email us at HaveYourSay@bbc.co.uk. Please include your name, age and location with any submission.
Read more →