Covid-19 Vaccine ‘Passports,’ Passes and Apps Around the Globe
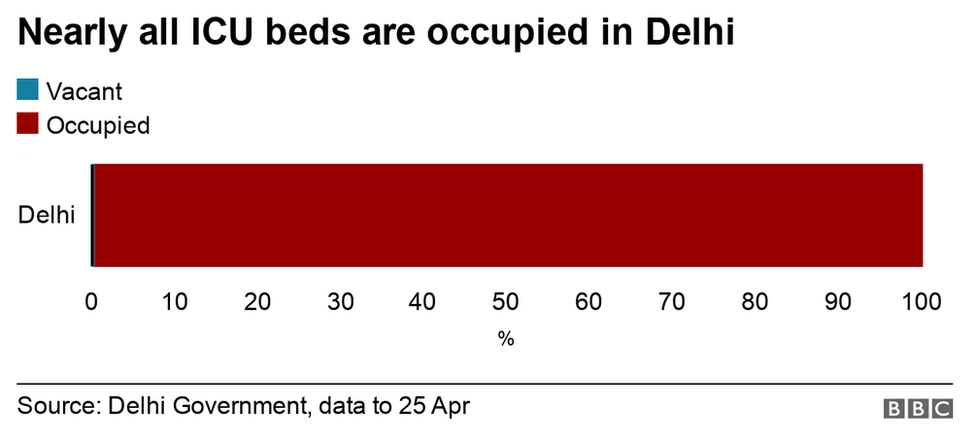
A variety of digital and old-fashioned approaches are being used around the globe to confirm Covid-19 vaccination. But the world is still far from a universal standard of proof.It is the latest status symbol. Flash it at the people, and you can get access to concerts, sports arenas or long-forbidden restaurant tables. Some day, it may even help you cross a border without having to quarantine.The new platinum card of the Covid age is the vaccine certificate. It is a document that has existed for more than two centuries, but it has rarely promised to hold so much power over culture and commerce. Many versions of these certificates now come with a digital twist.“It’s been a long time since we’ve had a pandemic that has impacted every facet of society so thoroughly, and then a vaccine,” said Carmel Shachar, the executive director of the Petrie-Flom Center for Health Law Policy, Biotechnology and Bioethics at Harvard Law School. “There is no precedent since 1918, and we definitely didn’t have smartphones in 1918.”Ramesh Raskar, a professor at M.I.T. Media Lab, has been leading an effort to develop a solution that includes both a paper certificate that anyone can easily carry as well as a free digital pass that works even without cell service.We are going to emerge from the pandemic with a new “currency for health,” he said.Figuring out how these passes should be used and what they should look like is dividing lawmakers, business leaders, ethicists, designers and health officials.Paper certificates from Shanghai, São Paulo and Uttar Pradesh, India. Anil Kumar Shukla and Ernesto Londoño/The New York Times An Easy Pass Made of Flimsy PaperA Covid-19 vaccination card does not always convey the significance of the document. Nor are the cards typically designed to counter fraud. Most of the hundreds of millions of people worldwide who got at least one shot of a Covid-19 vaccine in the past few months received a flimsy piece of paper.Unlike the easily recognizable “yellow card” that international travelers have long used to document other kinds of immunizations, the designs for Covid-19 vaccination certificates vary from state to state and country to country. In São Paulo, the cards have a green border. In Shanghai, they are stamped in red. In parts of Mexico and Lebanon, they are the size of passports, with the handwriting of the person who filled it out.In India, the certificate is a fully typed printed page. Next to the pronouncement that reads “Together, India will defeat Covid-19” is a photo of the country’s prime minister, Narendra Modi, a rare flourish.In some parts of the world, vaccine proof has gotten people a range of goods: free popcorn and ice cream, and even discounted beers. But for the most part, they have just allowed people to post selfies or to reassure their acquaintance. Some governments are looking for more formal systems that work on phones and counter fraud. Here is a look at some early efforts.DenmarkA template of Denmark’s vaccine passport.Denmark Ministry of HealthName: Coronapas (Corona Passport)Could it get you an indoor table? Yes. In Denmark, restaurants have been open for just takeout since December and have reopened this month with the caveat that only those with a Coronapas can sit indoors, the ministry of health said.How about a concert or sports game? That, too. As part of the country’s reopening plans, the government sanctioned indoor seating for sports and other stadium events for pass holders.Anything else? On April 6, hair salons, tattoo businesses, massage parlors and driving schools opened exclusively to customers with Coronapas. The Danish government does not track each time the pass is used, the ministry said, but about 3.5 million people had visited the app or site in its first week. The app is also intended to be a way for travelers to show other countries that they have been vaccinated.How do you get one? You have to be fully vaccinated and have tested negative for the coronavirus within the past 72 hours or overcame an infection within the past 180 days. Danish citizens can download the app on their smartphone or visit a website to print the Coronapas, which exudes the somber vibe of an old-school train ticket.Isn’t the European Union also developing a system? Yes. On June 21, the E.U. is expected to introduce a certificate called a Digital Green Pass, with the aim of allowing people who have been vaccinated against the coronavirus to travel more freely. Under the proposed rules, each nation within the bloc could decide which travel restrictions, such as obligatory quarantine, to waive for Digital Green holders. But many countries, including Denmark, say they cannot afford to wait for the Digital Green Pass and are developing their own versions.IsraelIsrael’s Green Pass app features a gif of a family headed on a trip.Name of card: The Green PassCould it get you an indoor table? Yes.How about entry to a concert or sports game? That, too.Anything else? The pass allows you to enter many businesses, including swimming pools, gyms, theaters and wedding halls, as well as cultural events, such as concerts, sports games and religious gatherings. Having the pass may also mean that you may not have to quarantine for 10 to 14 days after international travel.How does it work? In late February, Israel’s ministry of health began offering the Green Pass to fully vaccinated residents and individuals who have recovered from Covid-19. When booking a table at a restaurant, many of the businesses began to ask, “Do you have a Green Pass?” Israelis can print their certificates containing a QR code, download the code onto their phones or flash the app itself.What’s with that family? The app and other Green Pass materials feature an animated illustration of a family of three. The man is wearing shorts, a backpack and a camera around his neck, suggesting he’s on vacation. His son and wife are wearing masks, but their postures are relaxed as they pull their suitcases.Aparna Nair, a professor of science history at the University of Oklahoma who maintains a collection of vaccination certificates going back to the 1820s, said that this detail was noteworthy: “They are using the design of the vaccine passport to form visual connections with life after the pandemic, essentially, the vaccine as a literal passport to the rest of the world.”Are there concerns about requiring it so widely? Absolutely. Seema Mohapatra, a law professor specializing in health care and bioethics, noted that many people in the West Bank and Gaza strip had not been vaccinated, raising equity concerns.EstoniaA template of Estonia’s vaccine app.GuardtimeName of card: VaccineGuardCould it get you an indoor table? Not yet.How about entry to a concert or sports game? Not that either. Some business owners and cultural event organizers have expressed the hope that the Estonian government will waive capacity limitations and other restrictions if they agree to check for the certificate at the door, said Dr. Ain Aaviksoo, the chief medical officer at Guardtime, the company that worked with the Estonian government and the W.H.O. to design the certificate. But this hasn’t happened yet.So what is it good for? As of April 30, it will be most useful for going to and from Finland and back on the ferry.Is that really such a high-demand journey? Yes. About 10 percent of the Estonian working population is employed in Finland, Dr. Aaviksoo said. For years, it has been common for Estonians to stay in Helsinki during the week and then take the two-hour ferry ride home to Tallinn for the weekend. That has not been possible during the pandemic. Finland also fuels Estonia’s summer tourism.How does it work? Estonia says it cannot afford to wait for the E.U.’s Digital Green Pass. Starting April 30, residents can download a vaccination certificate, containing a QR code.New YorkA template of New York State’s Excelsior Pass which shows proof of vaccineNew York StateName: Excelsior PassCould it get you an indoor table? Yes.How about entry to a concert or sports game? That, too.Anything else? The state has been requiring some government employees to use it. It’s up to businesses and private organizations to decide whether they want to require the pass for entry. So far, the system has been used at restaurants, weddings, Madison Square Garden, Barclays Center and Yankee Stadium, according to Eric Piscini, vice president of Emerging Business Networks at IBM Watson Health, which designed it.How does it work? New York began offering downloads of the app in March. It verifies whether someone is fully vaccinated or has recently tested negative. Around 400,000 people throughout the state have downloaded it so far, Mr. Piscini said. The pass generates a QR code that can be scanned to produce a green checkmark or a red X. Those without compatible phones can print out their codes, he said.Who scans green? A person could obtain green not only by being fully vaccinated more than 14 days ago, but also by getting a negative PCR test within three days or a negative antigen test — often referred to as a rapid Covid test — within the past six hours. One challenge: only people vaccinated or tested in New York State can use it. “If you live in New Jersey, you will have to get tested in New York to get added to the database and go to the stadium,” Mr. Piscini said. IBM is trying to figure out how to access data from other states. Some venues may also accept other forms of certification.Will there ever be a national app for the U.S.? No. All of the 138 million Americans who have completed their first shot should have received the same Covid-19 vaccine record card created by the Centers for Disease Control and Prevention. But it’s up to states, universities and businesses to decide whether they want to require these cards or offer a supplemental app. The White House said in March there were no plans for a universal federal vaccination database or a mandate for a single credential. While New York has promoted Excelsior as the way to safely and quickly reopen the state, lawmakers in at least a half-dozen states, including Texas, Florida and Arkansas, have moved to ban businesses from requiring vaccination, saying it is a privacy violation or will slow down reopening of commerce.What differentiates Excelsior visually? It is the only Covid-19 vaccine pass to produce a large illustration of the Statue of Liberty when it is scanned.Isabel Kershner contributed reporting from Jerusalem, Suhasini Raj from New Delhi and Ernesto Londoño from Rio de Janeiro
Read more →